Python API¶
This section includes information for using the pure Python API of bob.learn.libsvm.
- bob.learn.libsvm.get_include()[source]¶
Returns the directory containing the C/C++ API include directives
- class bob.learn.libsvm.File¶
Bases: object
File(path)
Loads a given LIBSVM data file. The data file format, as defined on the library README is like this:
<label> <index1>:<value1> <index2>:<value2> ... <label> <index1>:<value1> <index2>:<value2> ... <label> <index1>:<value1> <index2>:<value2> ... ...
The labels are integer values, so are the indexes, starting from 1 (and not from zero as a C-programmer would expect). The values are floating point. Zero values are suppressed - LIBSVM uses a sparse format.
Upon construction, objects of this class will inspect the input file so that the maximum sample size is computed. Once that job is performed, you can read the data in your own pace using the File.read() method.
This class is made available to you so you can input original LIBSVM files and convert them to another better supported representation. You cannot, from this object, save data or extend the current set.
- eof() → bool¶
Returns True if the file has reached its end. To start reading from the file again, you must call File.reset() before another read operation may succeed.
- fail() → bool¶
Returns True if the file has a fail condition or bad bit sets. It means the read operation has found a critical condition and you can no longer proceed in reading from the file. Note this is not the same as File.eof() which informs if the file has ended, but no errors were found during the read operations.
- filename¶
The name of the file being read
- good() → bool¶
Returns if the file is in a good state for readout. It is True if the current file it has neither the eof, fail or bad bits set, whic means that next File.read() operation may succeed.
- read([values]) -> (int, array)¶
Reads a single line from the file and returns a tuple containing the label and a numpy array of float64 elements. The numpy.ndarray has a shape as defined by the File.shape attribute of the current file. If the file has finished, this method returns None instead.
If the output array values is provided, it must be a 64-bit float array with a shape matching the file shape as defined by File.shape. Providing an output array avoids constant memory re-allocation.
- read_all([labels, [values]) -> (array, array)¶
Reads all contents of the file into the output arrays labels (used for storing each entry’s label) and values (used to store each entry’s features). The array labels, if provided, must be a 1D numpy.ndarray with data type int64, containing as many positions as entries in the file, as returned by the attribute File.samples. The array values, if provided, must be a 2D array with data type float64, as many rows as entries in the file and as many columns as features in each entry, as defined by the attribute File.shape.
If the output arrays labels and/or values are not provided, they will be allocated internally and returned.
Note
This method is intended to be used for reading the whole contents of the input file. The file will be reset as by calling File.reset() before the readout starts.
- reset() → None¶
Resets the current file so it starts reading from the begin once more.
- samples¶
The number of samples in the file
- shape¶
The size of each sample in the file, as tuple with a single entry
- class bob.learn.libsvm.Machine¶
Bases: object
Machine(path)
Machine(hdf5file)
This class can load and run an SVM generated by libsvm. Libsvm is a simple, easy-to-use, and efficient software for SVM classification and regression. It solves C-SVM classification, nu-SVM classification, one-class-SVM, epsilon-SVM regression, and nu-SVM regression. It also provides an automatic model selection tool for C-SVM classification. More information about libsvm can be found on its website. In particular, this class covers most of the functionality provided by the command-line utility svm-predict.
Input and output is always performed on 1D or 2D arrays with 64-bit floating point numbers.
This machine can be initialized in two ways: the first is using an original SVM text file as produced by libsvm. The second option is to pass a pre-opened HDF5 file pointing to the machine information to be loaded in memory.
Using the first constructor, we build a new machine from a libsvm model file. When you load using the libsvm model loader, note that the scaling parameters will be set to defaults (subtraction of 0.0 and division by 1.0). If you need scaling to be applied, set it individually using the appropriate methods on the returned object.
Using the second constructor, we build a new machine from an HDF5 file containing not only the machine support vectors, but also the scaling factors. Using this constructor assures a 100% state recovery from previous sessions.
- coef0¶
The coefficient 0 for 'POLY' (polynomial) or 'SIGMOIDAL' (sigmoidal) kernels
- degree¶
The polinomial degree, only valid if the kernel is 'POLY' (polynomial)
- forward(input[, output]) → array¶
o.predict_class(input, [output]) -> array
o(input, [output]) -> array
Calculates the predicted class using this Machine, given one single feature vector or multiple ones.
The input array can be either 1D or 2D 64-bit float arrays. The output array, if provided, must be of type int64, always uni-dimensional. The output corresponds to the predicted classes for each of the input rows.
Note
This method only accepts 64-bit float arrays as input and 64-bit integers as output.
- gamma¶
The
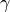 parameter for 'POLY' (polynomial),
'RBF' (gaussian) or 'SIGMOID' (sigmoidal) kernels
parameter for 'POLY' (polynomial),
'RBF' (gaussian) or 'SIGMOID' (sigmoidal) kernels
- input_divide¶
Input division factor, before feeding data through the weight matrix W. The division is applied just after subtraction - by default, it is set to 1.0.
- input_subtract¶
Input subtraction factor, before feeding data through the weight matrix W. The subtraction is the first applied operation in the processing chain - by default, it is set to 0.0.
- kernel_type¶
The type of kernel used by the support vectors in this machine
- labels¶
The class labels this machine will output
- machine_type¶
The type of SVM machine contained
- predict_class()¶
o.forward(input, [output]) -> array
o.predict_class(input, [output]) -> array
o(input, [output]) -> array
Calculates the predicted class using this Machine, given one single feature vector or multiple ones.
The input array can be either 1D or 2D 64-bit float arrays. The output array, if provided, must be of type int64, always uni-dimensional. The output corresponds to the predicted classes for each of the input rows.
Note
This method only accepts 64-bit float arrays as input and 64-bit integers as output.
- predict_class_and_probabilities(input, [cls, [prob]]) -> (array, array)¶
Calculates the predicted class and output probabilities for the SVM using the this Machine, given one single feature vector or multiple ones.
The input array can be either 1D or 2D 64-bit float arrays. The cls array, if provided, must be of type int64, always uni-dimensional. The cls output corresponds to the predicted classes for each of the input rows. The prob array, if provided, must be of type float64 (like input) and have as many rows as input and len(o.labels) columns, matching the number of classes for this SVM.
This method always returns a tuple composed of the predicted classes for each row in the input array, with data type int64 and of probabilities for each output of the SVM in a 1D or 2D float64 array. If you don’t provide the arrays upon calling this method, we will allocate new ones internally and return them. If you are calling this method on a tight loop, it is recommended you pass the cls and prob arrays to avoid constant re-allocation.
- predict_class_and_scores(input, [cls, [score]]) -> (array, array)¶
Calculates the predicted class and output scores for the SVM using the this Machine, given one single feature vector or multiple ones.
The input array can be either 1D or 2D 64-bit float arrays. The cls array, if provided, must be of type int64, always uni-dimensional. The cls output corresponds to the predicted classes for each of the input rows. The score array, if provided, must be of type float64 (like input) and have as many rows as input and C columns, matching the number of combinations of the outputs 2-by-2. To score, LIBSVM will compare the SV outputs for each set two classes in the machine and output 1 score. If there is only 1 output, then the problem is binary and only 1 score is produced (C = 1). If the SVM is multi-class, then the number of combinations C is the total amount of output combinations which is possible. If N is the number of classes in this SVM, then
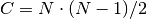 .
If N = 3, then C = 3. If N = 5, then C = 10.
.
If N = 3, then C = 3. If N = 5, then C = 10.This method always returns a tuple composed of the predicted classes for each row in the input array, with data type int64 and of scores for each output of the SVM in a 1D or 2D float64 array. If you don’t provide the arrays upon calling this method, we will allocate new ones internally and return them. If you are calling this method on a tight loop, it is recommended you pass the cls and score arrays to avoid constant re-allocation.
- probability¶
Set to True if this machine supports probability outputs
- save(path) → None¶
o.save(hdf5file) -> None
Saves itself at a LIBSVM model file or into a bob.io.HDF5File. Saving the SVM into an bob.io.HDF5File object, has the advantage of saving input normalization options together with the machine, which are automatically reloaded when you re-initialize it from the same HDF5File.
- shape¶
A tuple that represents the size of the input vector followed by the size of the output vector in the format (input, output).
- class bob.learn.libsvm.Trainer¶
Bases: object
Trainer([machine_type=’C_SVC’, [kernel_type=’RBF’, [cache_size=100, [stop_epsilon=1e-3, [shrinking=True, [probability=False]]]]]]) -> new Trainer
This class emulates the behavior of the command line utility called svm-train, from LIBSVM. It allows you to create a parameterized LIBSVM trainer to fullfil a variety of needs and configurations. The constructor includes parameters which are global to all machine and kernel types. Specific parameters for specific machines or kernel types can be fine-tuned using object attributes (see help documentation).
Parameters:
- machine_type, str
The type of SVM to be trained. Valid options are:
- 'C_SVC' (the default)
- 'NU_SVC'
- 'ONE_CLASS' (unsupported)
- 'EPSILON_SVR' (unsupported regression)
- 'NU_SVR' (unsupported regression)
- kernel_type, str
The type of kernel to deploy on this machine. Valid options are:
- 'LINEAR', for a linear kernel
- 'POLY', for a polynomial kernel
- 'RBF', for a radial-basis function kernel
- 'SIGMOID', for a sigmoidal kernel
- 'PRECOMPUTED', for a precomputed, user provided kernel please note this option is currently unsupported.
- cache_size, float
- The size of LIBSVM’s internal cache, in megabytes
- stop_epsilon, float
- The epsilon value for the training stopping criterion
- shrinking, bool
- If set to True (the default), the applies LIBSVM’s shrinking heuristic.
- probability, bool
- If set to True, then allows the outcoming machine produced by this trainer to output probabilities besides scores and class estimates. The default for this option is False.
Note
These bindings do not support:
- Precomputed Kernels
- Regression Problems
- Different weights for every label (-wi option in svm-train)
Fell free to implement those and remove these remarks.
- cache_size¶
Internal cache size to be used by LIBSVM (in megabytes)
- coef0¶
The coefficient 0 for 'POLY' (polynomial) or 'SIGMOID' (sigmoidal) kernels
- cost¶
The cost value for C_SVC, EPSILON_SVR or NU_SVR. This parameter is normally referred only as
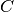 on
literature. It should be a non-negative floating-point number.
on
literature. It should be a non-negative floating-point number.
- degree¶
The polinomial degree, only used if the kernel is 'POLY' (polynomial)
- gamma¶
The
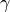 parameter for 'POLY' (polynomial),
'RBF' (gaussian) or 'SIGMOID' (sigmoidal) kernels
parameter for 'POLY' (polynomial),
'RBF' (gaussian) or 'SIGMOID' (sigmoidal) kernels
- kernel_type¶
The type of kernel used by the support vectors in this machine
- loss_epsilon_svr¶
For EPSILON_SVR, this is the
 value
on the equation
value
on the equation
- machine_type¶
The type of SVM machine that will be trained
- nu¶
The nu value for NU_SVC, ONE_CLASS or NU_SVR. This parameter should live in the range [0, 1].
- probability¶
If set to True, output Machines will support outputting probability estimates
- shrinking¶
If set to True, then use LIBSVM’s Shrinking Heuristics
- stop_epsilon¶
The epsilon used for stop training
- train(data[, subtract, divide]) → array¶
Trains a new machine for multi-class classification. If the number of classes in data is 2, then the assigned labels will be +1 and -1, in that order. If the number of classes is greater than 2, labels are picked starting from 1 (i.e., 1, 2, 3, 4, etc.). This convention follows what is done at the command-line for LIBSVM.
The input object data must be an iterable object (such as a Python list or tuple) containing 2D 64-bit float arrays each representing data for one single class. The data in each array should be organized row-wise (i.e. 1 row represents 1 sample). All rows for all arrays should have exactly the same number of columns - this will be checked.
Optionally, you may also provide both input arrays subtract and divide, which will be used to normalize the input data before it is fed into the training code. If provided, both arrays should be 1D and contain 64-bit floats with the same width as all data in the input array data. The normalization is applied in the following way: